Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Genotypic Strains Similarity of Mycobacterium Tuberculosis Complex from Pulmonary Patients in North-Central Nigeria
*Corresponding author: Ajide BA, Bingham University, Karu, Nasarawa, Nigeria.
Received: October 24, 2023; Published: November 08, 2024
DOI: 10.34297/AJBSR.2024.24.003235
Abstract
Nigeria has a high rate of Tuberculosis (TB) and a high burden of TB/HIV infections. To enhance TB control in Nigeria, an understanding of the genotyping and drug resistance of Mycobacterium Tuberculosis Complex (MTBC) is critical. This study investigated the genetic diversity of MTBC strains that are associated with pulmonary tuberculosis in North-Central Nigeria. Sputum samples of 2800 suspected Pulmonary Tuberculosis (PTB) patients were collected and 371 positive cases cultured using Lowenstein-Jensen medium. The isolates’ DNA was extracted and then subjected to region of difference analysis and spoligotyping. Biomimetics software was used to build the phylogenetic tree to compare isolates with similar genetic patterns. A total of 272 DNA samples were subjected to spoligotyping to build a phylogenetic tree. About 50.37% were defined and registered in the SIT database, with Cameroon (CAM) having the highest number 34.008% of isolates followed by Beijing and H1 lineages with 6.24% and 4.76% respectively. Exactly 124 patterns were found with 16.94% identified and 83.07% undefined. The distribution of new/orphan isolates included two identified lineages without SIT numbers: AFRI_3 (1.10%) and T1 (0.37%). The defined patterns 50.37% were matched in the SITVIT2 databases, and 11 strains were clustered with 92.70% of isolates, while 7.29% were distinct strains, resulting in a 45.59% overall strain diversity. SIT (2832), SIT (61), and SIT (1) were the most common strains found with 23.08%, 7.69% and 4.40% isolates, respectively. The origin and geographic distributions of different lineages were also confirmed to be geographically dispersed. Consequently, with the discovery of 103 new strains, this study has provided insight into the genetic diversity of MTBC and types of strains circulating in the study area, providing a platform for public health information gathering and further investigation.
Keywords: Genotyping, Drug resistance, Mycobacterium tuberculosis complex, Pulmonary patients, Phylogenetic tree, Spoligotypin
Introduction
Tuberculosis (TB) remains the most contagious disease in humans with one of the highest mortality rates worldwide [1-3]. It is a granulomatous inflammatory infection mainly caused by Mycobacterium tuberculosis and the classical Mycobacterium tuberculosis complex (MTBC); affecting the lungs, central nervous system, lymphatic and circulatory system [4,5]. M. tuberculosis affects one-third of the world’s population and is the second leading cause of death after the human immunodeficiency virus (HIV) [6].
Despite the availability of vaccination and preventive screening programs such as the Bacillus Calmette Guerin (BCG) vaccine [7] as well as curative treatments with about 90% case efficacy, TB remains a major global health challenge with a global prevalence of 10 million people (1.1 million in children) and a global mortality rate of 1.5 million people (251,000 in children) in 2020 [8]. Rifampicin is the first-line treatment for TB. However, an incidence of 484,000 Rifampicin-resistant cases was reported in 2020 with 78% multi-drug-resistant (MDR-TB) cases [8]. Treatment of TB is a difficult task which requires administration of multiple antibiotics over a long period. Poor management of TB care; including incorrect drug prescribing practices, poor quality of drugs and patient nonadherence are the major causes of parasite resistance to available TB drugs. Although it is estimated that 60 million lives were saved through TB diagnosis and treatment between 2000 and 2020 [8] and the global TB incidence is falling at about 2% per annum, yet the 2020 target for the End TB Strategy; which was one of the health targets of the Sustainable Development Goals remains unmet.
Nigeria is one of the countries with a high prevalence of tuberculosis, tuberculosis/HIV, and MDR-TB. The country is ranked seventh of the 30 countries with the highest prevalence of Tuberculosis (TB) worldwide, and second in Africa [8] Nigeria's tuberculosis crisis is further exacerbated by the phenomenon of drug-resistance as well as the HIV/AIDS epidemic. Since HIV raises the risk of TB progression [9], the risk factors associated with HIV in the population must be considered for successful control [10]. An estimated 407,000 HIV-negative people are believed to be infected with tuberculosis in Nigeria annually. The North-Central Region, which comprises six states and the Federal Capital Territory (FCT), has the highest HIV prevalence of Nigeria's six geopolitical zones [11] and an estimated 63,000 HIV-positive individuals contract tuberculosis in this region per year [11]. No factors influencing the high prevalence of HIV and associated diseases in this geopolitical region is reported, but extensive socio-epidemiological and behavioural research have been suggested.
Regrettably, of the 407,000 annual projected tuberculosis cases, only 104,904 tuberculosis cases were diagnosed in 2017. This is indicative of a poor TB case finding rate in both adults and children and equates to a treatment coverage of just 25.8%. This is a major problem in Nigeria as it suggests that nearly 75% (302,096) of people with tuberculosis are either undetected or detected but not notified, especially in "non-DOTS sites" [12] Hence, instituting effective TB surveillance programs with capacity for prompt new case diagnosis, appropriate therapy provision, and accurate detection of outbreaks in communities, in order to implement proper TB control and elimination strategies is imperative [13,14]. To achieve this goal, information from classical and molecular epidemiology needs to be combined with patient clinical data. Thus, identification of strains circulating in a certain geographical region using molecular tools can contribute to tracking transmission and thus improving the TB control program of that region. The importance of genetic diversity studies as fundamental tools for gaining insight into a population, species or subspecies is well documented [15-18]. This study aims to profile the genetic diversity or relatedness of Mycobacterium tuberculosis complex isolates from Pulmonary Tuberculosis (PTB) patients in the north central of Nigeria.
Study Area and Study Population
This study was carried out across fourteen general hospitals (2 each in the local government) in the seven states of the North Central Geopolitical Zone of Nigeria. The North Central region is geographically located in Nigeria's middle belt region, stretching from west to east along the confluence of the Niger and Benue rivers. Niger, Kogi, Benue, Plateau, Nassarawa, Kwara, and the Federal Capital Territory of Abuja make up the area. The presence of improved diagnostic services, Directly Observed Treatment Short-course (TB-DOTS) centres, professional human resources as well as the high prevalence of HIV, were the primary reasons for choosing this region.
Some 371 positive sputa samples were collected from 2800 suspected pulmonary TB patients in this chosen area between 2017 and 2018 using a basic random sampling process [19] (Figure 1).
Molecular Identification
A total of 272 positive culture colonies drawn from 371 positive sputa were extracted from the LJ media surface and their DNA extracted using Geno-Lyse kit (Hain Life-science GmbH, Nehren, Germany) based on manufacturer’s instructions. The extracted DNA was used for spoligotyping at the DST/NRF Centre of Excellence for Biomedical TB Research (CBTBR), National Health Laboratory Service (NHLS) Johannesburg South Africa. All protocols used for the spoligotyping of the 272 DNA isolates were according to laboratory SOP.
Interpretation of the Results
Analysis of the results of the spoligotype membrane autoradiographs was based on the appearance or absence of signals at the sites of DNA/DNA hybridization. On video, the existence of spacers was depicted as black squares. The results were entered into spreadsheets in binary format and linked to the publicly available spoligotyping database SpolDB4 (http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/).
Result
Out of 272 DNA isolates from pure culture of MTBC that were spoligotyped, 137 (50.37%) were identified and registered in the SIT database. The Cameroon Lineage (CAM) had the highest number of isolates with 93 (34.08 %), followed by Beijing and H1 types with 17 (6.24 %) and 13 (4.76 %), respectively. Notably, there were 135 (49.63 %) new and orphan isolates (Figure 2).
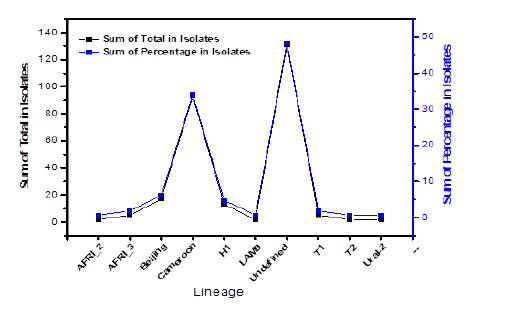
Figure 2: Sum and percentages of all spoligotyped MTB isolated from pulmonary patients across North central Nigeria.
There were 124 patterns recorded in total, with 21 (16.94%) identified patterns and 103 (83.07%) unknown patterns. Figure 3 depicts the distribution of new/orphan (131; 48.25%) isolates including two distinct lineages with SIT numbers; AFRI_3 3 (1.10%) and T1 1 (0.37%).
Table 1 shows that all 21 of the trends fit in the SITVIT2 or SpolDB4 databases, with 11 strains accounting for 127 (92.70%) isolates and 10 strains accounting for 7.29% of the total, resulting in a 45.59 % average variety of the strains. SIT 2832, SIT 61, and SIT 1 were the most common strains found, with 63, 21, and 12 isolates, respectively (Table 1). Origin and geographic distributions of various lineages were also confirmed to be geographically spread in this study with their SIT across regions (Table 2).
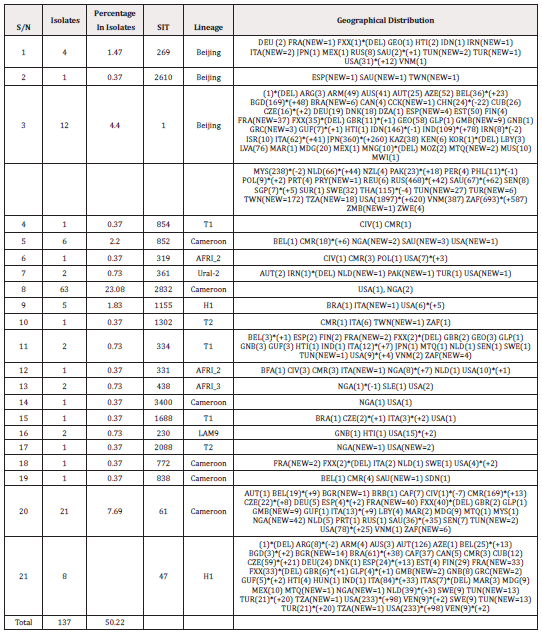
Table 1: Twenty-one spoligotyped isolates from North central Nigeria registered in the data base database.
Phylogenetic tree was designed using Biomimetics software to evaluate isolates with comparable genetic patterns (Figure 4).
Discussion
Despite being treatable, TB is a major global health issue, claiming the lives of over two million people per year. The genetic diversity of M. tuberculosis complex strains circulating in North Central Nigeria and involved in Pulmonary Tuberculosis (PTB) was studied in this research. While M. tuberculosis strain variability has been recorded in some parts of Nigeria, the current study found that strains belonging to the CAM (Cameroon) lineage are the most prevalent in North Central Nigeria.
In this analysis, a total of 272 culture isolates were spoligotyped, yielding 124 patterns, 21 (16.13%) of which were identified and reported in the SIT data base, while 103 (83.07%) were not defined, i.e., did not fit any pattern in the SIT data base. A total of 135 (49.63%) were found to be entirely new or orphan, as well as not identified in the data base, implying that they had never been documented before. Two of these, AFRI_3 3 (1.10%) and T1 1 (0.37%), indicated their lineage but were new/orphan, and 137 (50.37%) were defined. The high number of undefined strains may be because not many genetic diversity studies of MTBC have been performed in this geographical field (Table 2). The origins of the prevalent linages and other shared forms are known in terms of spatial distribution, while the origins of others are unknown [21].
The CAM lineage, which has phylogeographical specificity for Cameroon and neighbouring West African nations, accounted for the majority of isolates, with 93 (34.08%), followed by Beijing 17 (6.24%) and H1 13 (4.76%). CAM is the most predominant lineage circulating across the North Central Nigeria and this corroborates the work of [22], who reported a prevalence of 66% for CAM clade in their study [23]. Also reported the CAM lineage as the predominant type (76%) in Jos, a major city in North Central Nigeria while [24] reported CAM to have the highest prevalence of 49.44% in a study on the genetic variation of MTBC in Southeast Nigeria.
Similar studies in other neighbouring countries: Chad, Sierra Leone and Burkina Faso have also established CAM as the predominant strain isolated in PTB patients with the LAM 10-CAM sub-lineage being the predominantly spoligotyped sub lineage in neighbouring countries of Cameroon and Benin republic [25-28]. The LAM 10-CAM sub-lineage is known to have originated in Cameroon [29]. The fact that this genotype MTBC strain is characteristic of the Western and Central Africa region may be the explanation for the highest frequency of CAM reported in our sample.
This may be explained by trading interactions bringing about transmission. Cameroon for example shares a border with Benue state in North Central Nigeria, where the pressure is dominant. The Benue province being an agricultural hub draws considerable trading ties, migration for farming purposes as well as inter-cultural marriages. The Bakassi Peninsula for example is today a province of Cameroon despite being for many decades heavily inhabited by Nigerian natives. This may help disperse CAM tensions from Cameroon to Nigeria's north central region and further coupled with the high TB/HIV burden that has always been recorded in Benue state could explain the predominant genotyped strain identified in this sample. Other countries like South Africa, Czech Republic, Russia, India have also been identified as evolutionary phylogenetic places of origin for different sero-type strains of CAM [21,29].
The Beijing family is considered to have originated from China where it was first identified in 1995 and termed Beijing because a lot of the strains were isolated from TB patients residing in Beijing [30]. The Beijing genotype is known to have circulated in Beijing's environs from 1956 to 1960 before spreading to neighbouring Asian countries such as Mongolia, Thailand, South Korea, Vietnam and further to other regions of the world [31].
Although the AFRI_2 and 3 lineages are also thought to play a role in human tuberculosis, the family is still poorly known. In terms of origin, they are considered more unique to Russia despite being found all over the world. Guinea-Bissau, Gambia, Sierra Leone, Senegal, Burkina Faso, Cameroon, Nigeria, and Côte d'Ivoire are among the west African countries where the AFRI 2 and 3 lineages have been found [26,32-36] while the LAM 9 was discovered in South Africa and Somalia for the first time.
When the spoligotyped isolates in this research area were compared to international records, all 21 patterns aligned with the SITVIT2 or SpolDB4 databases, and out of these patterns, 127 (92.70 %) isolates were grouped, while 10 strains were found to be distinct (7.29%), giving the strains an overall diversity of 45.59%. SIT 2832/n = 63 (23.08 %), SIT 1/n = 12 (4.40%), and SIT 61/n = 21 are the three most common strains isolated (7.69 %) (Table 1). This reveals that they are in circulation outside of north central Nigeria, in other geographical zones, and across the globe, which is consistent with the findings of previous analysis in Nigeria [22,37]. This indicates that, the lineage contributes in no small part to the transmission of TB in North Central Nigeria, and there may be ongoing spread of PTB according to a cluster rate of 45.59% found in this study area. This should however be taken with caution because spoligotyping, when used alone, has lower discriminatory capacity than when combined with other genotyping approaches.
Furthermore, phylogenetic tree data (Figure 4) which evaluated and grouped isolates with similar genetic characteristics, indicated that the CAM lineage strains were the most prevalent cluster, with the largest strains, and hance are most likely responsible for infection transmission within the population [38].
Conclusion
The current research has shed light on Mycobacterium's genetic diversity. Sputum from alleged PTB patients in North Central Nigeria yielded tuberculosis complex isolates with CAM as the prevailing lineage. The dominance of this lineage in the study region suggests that it will become more common across the country in the near future. Its similarity and affiliation in the international data bank also indicates its predominant role in triggering the PTB outbreak in North Central Nigeria. Importantly, data from this work provides a platform of information that can be synergized with observations from other geographical zones in recognizing risk disparities to help prioritize resource allocation for TB surveillance, monitoring, and prevention. The database would almost certainly include the latest/orphan and undefined strains found in this analysis, but those isolates might not be new for very long.
Acknowledgments
None.
Conflict of Interest
None.
References
- Cattamanchi A (2020) Tuberculosis (TB): Causes, symptoms, and treatments [WWW Document].
- (2017) The Conversation Four of the most lethal infectious diseases of our time and how we’re overcoming them [WWW Document].
- World Health Organization (2018) WHO - The top 10 causes of death [WWW Document]. 24 Maggio. URL http://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death
- Golden M P, Vikram H R (2005) Extrapulmonary tuberculosis: An overview. Am Fam Physician 72(9): 1761-1768.
- Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H, et al. (2010) Mycobacterial outer membranes: in search of proteins. Trends Microbiol 18(3): 109-116.
- Fitzgerald D W, Sterling T R, Haas D W (2014) Mycobacterium tuberculosis, in: Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Elsevier Inc: 2787.
- Gantz N M (2001) Management of Antimicrobials in Infectious Diseases: Impact of Antibiotic Resistance, in: Annals of Inter Med Humana Press, New York, 144.
- World Health Organization (2021) Global Tuberculosis Report, Blood. Geneva. https://doi.org/978 92 4 156450 2
- Obasanya J, Abdurrahman ST, Oladimeji O, Lawson L, Dacombe R, et al. (2015) Tuberculosis case detection in Nigeria, the unfinished agenda. Trop Med Int Heal 20(10): 1396-1402.
- Dandona L, Dandona R, Kumar G A, Reddy G B, Ameer M A, Ahmed G M, et al. (2008) Risk factors associated with HIV in a population-based study in Andhra Pradesh state of India. Int J Epidemiol 37: 1274-1286.
- Federal Ministry of Health, 2015. 2014 National HIV Sero-prevalence Sentinel Survey among Pregnant Women Attending Antenatal Clinics in Nigeria: Technical Report, national AIDS/STI Control Programme.
- Kanabus A (2020) TB in Nigeria - Funding, children, diagnosing TB, HIV/TB [WWW Document]. GHE Inf about Tuberc.
- Lienhardt C, Glaziou P, Uplekar M, Lå ̈nnroth K, Getahun H, et al. (2012) Global tuberculosis control Lessons learnt and future prospects. Nat Rev Microbiol 10(6): 407-416.
- Sandgren A, Schepisi MS, Sotgiu G, Huitric E, Migliori GB, et al. (2014) Tuberculosis transmission between foreign- and native-born populations in the EU/EEA: A systematic review. in European Respiratory Journal European Respiratory Society 43(3): 1159-1171.
- Avolio M L, Beaulieu J M, Lo E Y Y, Smith M D (2012) Measuring genetic diversity in ecological studies. Plant Ecol 213: 1105-1115.
- Mukhopadhyay T, Bhattacharjee S (2016) Genetic Diversity: Its Importance and Measurements.
- Sheriff O, Alemayehu K (2018) Genetic diversity studies using microsatellite markers and their contribution in supporting sustainable sheep breeding programs: A review. Cogent Food Agric 4.
- Zhang C, Jia C, Liu X, Zhao H, Hou L, Li M, et al. (2022) Genetic Diversity Study on Geographical Populations of the Multipurpose Species Elsholtzia stauntonii Using Transferable Microsatellite Markers. Front Plant Sci 13: 1-14.
- Van Der Werf M J, Borgdorff M W (2007) How to measure the prevalence of tuberculosis in a population. Trop Med Int Heal 12(4): 475-484.
- Ideki O, Weli V E (2019) Assessment of Drought Vulnerability and Occurrence Zones in North Central Nigeria. Atmos Clim Sci 9: 298-309.
- Warren R M, Streicher E M, Sampso S L, Van der Spuy G D, Richardson M, Nguyen D, et al. (2002) Microevolution of the direct repeat region of Mycobacterium tuberculosis: Implications for interpretation of spoligotyping data. J Clin Microbiol 40(12): 4457-4465.
- Uzoewulu G N, Lawson L, Nnanna I S, Rastogi N, Goyal M (2016) Genetic diversity of Mycobacterium tuberculosis complex strains isolated from patients with pulmonary tuberculosis in Anambra State, Nigeria. Int J Mycobacteriol 5(1): 74-79.
- Ani A, Bruvik T, Okoh Y, Agaba P, Agbaji O, Idoko J, et al. (2010) Genetic diversity of Mycobacterium tuberculosis Complex in Jos, Nigeria. BMC Infect Dis 26(10): 189.
- Nuru A, Mamo G, Worku A, Admasu A, Medhin G, et al. (2015) Genetic Diversity of Mycobacterium tuberculosis Complex Isolated from Tuberculosis Patients in Bahir Dar City and Its Surroundings, Northwest Ethiopia. Biomed Res. Int.
- Godreuil S, Torrea G, Terru D, Chevenet F, Diagbouga S, Supply P, et al. (2007) First molecular epidemiology study of Mycobacterium tuberculosis in Burkina Faso. J Clin Microbiol 45(3): 921-927.
- Homolka S, Post E, Oberhauser B, George A G, Westman L, Dafae F, et al. (2008) High genetic diversity among Mycobacterium tuberculosis complex strains from Sierra Leone. BMC Microbiol 8: 103.
- Panwalkar N, Chauhan DS, Desikan P (2017) Spoligotype defined lineages of Mycobacterium tuberculosis and drug resistance: Merely a casual correlation? Indian J Med Microbiol 35(1): 27-32.
- Yeboah Manu D, Asante Poku A, Bodmer T, Stucki D, Koram K, Bonsu F, et al. (2011) Genotypic diversity and drug susceptibility patterns among M. tuberculosis complex isolates from South-Western Ghana. PLoS One 6(7): 21906.
- Niobe Eyangoh SN, Kuaban C, Sorlin P, Cunin P, Thonnon J, et al. (2003) Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 41(6): 2547-2553.
- Van Soolingen D (1998) Utility of molecular epidemiology of tuberculosis. Eur Respir J 11(4): 795-797.
- Toungoussova OS, Mariandyshev A, Bjune G, Sandven P, Caugant DA, et al. (2003) Molecular epidemiology and drug resistance of Mycobacterium tuberculosis isolates in the Archangel prison in Russia: Predominance of the W-Beijing clone family. Clin. Infect. Dis. 37, 665–672.
- Cadmus S, Palmer S, Okker M, Dale J, Gover K, Smith N, et al. (2006) Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J Clin Microbiol 44(1): 29-34.
- Källenius G, Koivula T, Ghebremichael S, Hoffner S E, Norberg R, Svensson E, er al. (1999) Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J Clin Microbiol 37(12): 3872-3878.
- Ledru S, Cauchoix B, Yaméogo M, Zoubga A, Lamandé Chiron, et al. (1996) Impact of short-course therapy on tuberculosis drug resistance in South-West Burkina Faso. Tuber Lung Dis 77(5): 429-436.
- Mahamar A, Andemel N, Swihart B, Sidibe Y, Gaoussou S, et al. (2021) Malaria infection is common and associated with perinatal mortality and preterm delivery despite widespread use of chemoprevention in Mali: an observational study 2010 to 2014. Clin Infect Dis 73(8): 1355-1361.
- Niemann S, Kubica T, Bange FC, Adjei O, Browne EN, et al. (2004) The species Mycobacterium africanum in the light of new molecular markers. J Clin Microbiol 42(9): 3958-3962.
- Ravansalar H, Tadayon K, Mosavari N, Derakhshan M, Ghazvini K, et al. (2017) Genetic diversity of Mycobacterium tuberculosis complex isolated from patients in the Northeast of Iran by MIRU-VNTR and spoligotyping. Jundishapur J Microbiol 10.
- Guernier V, Sola C, Brudey K, Guégan J F, Rastogi N (2008) Use of cluster-graphs from spoligotyping data to study genotype similarities and a comparison of three indices to quantify recent tuberculosis transmission among culture positive cases in French Guiana during a eight year period. BMC Infect Dis 8: 46.







 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.